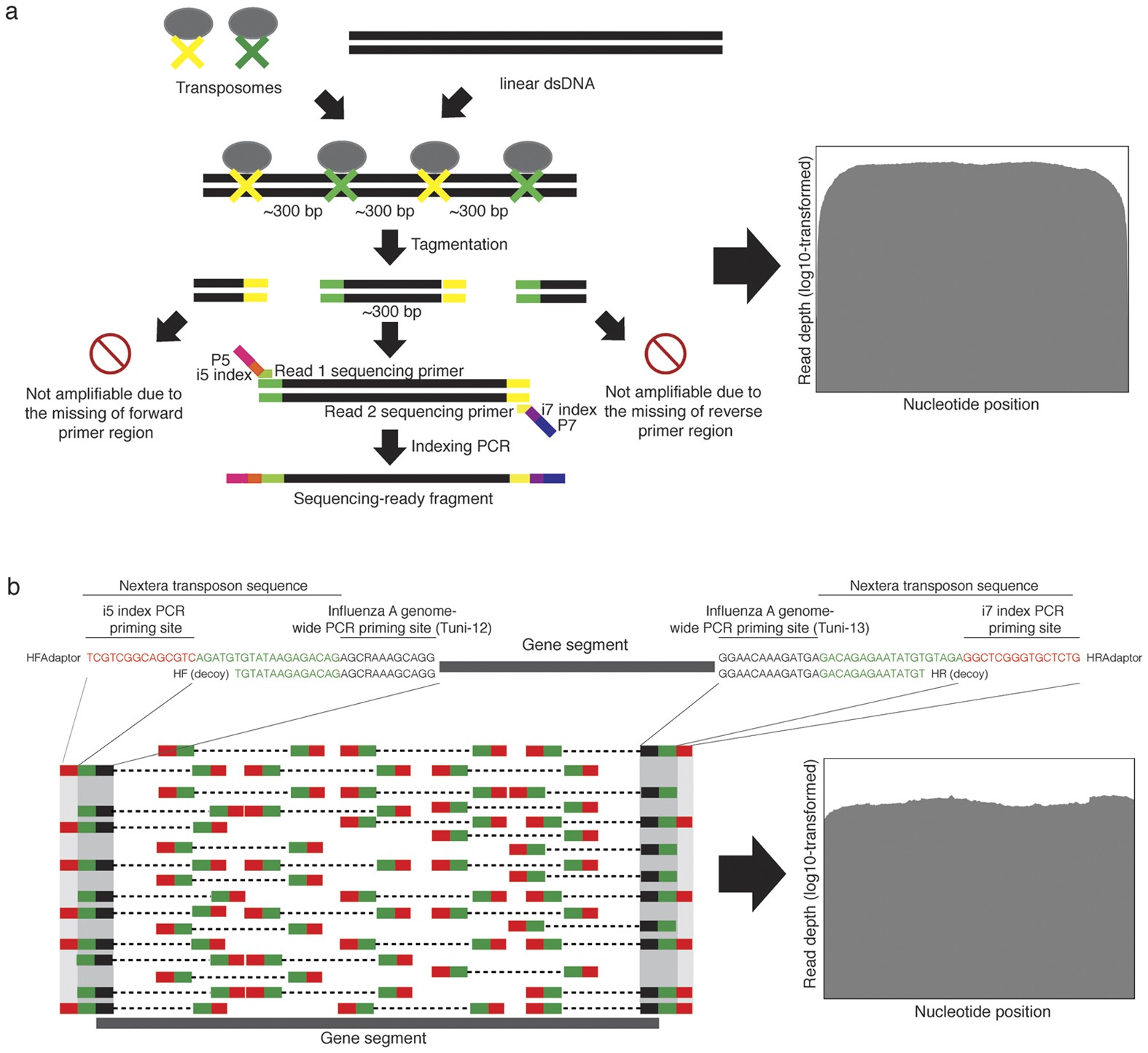
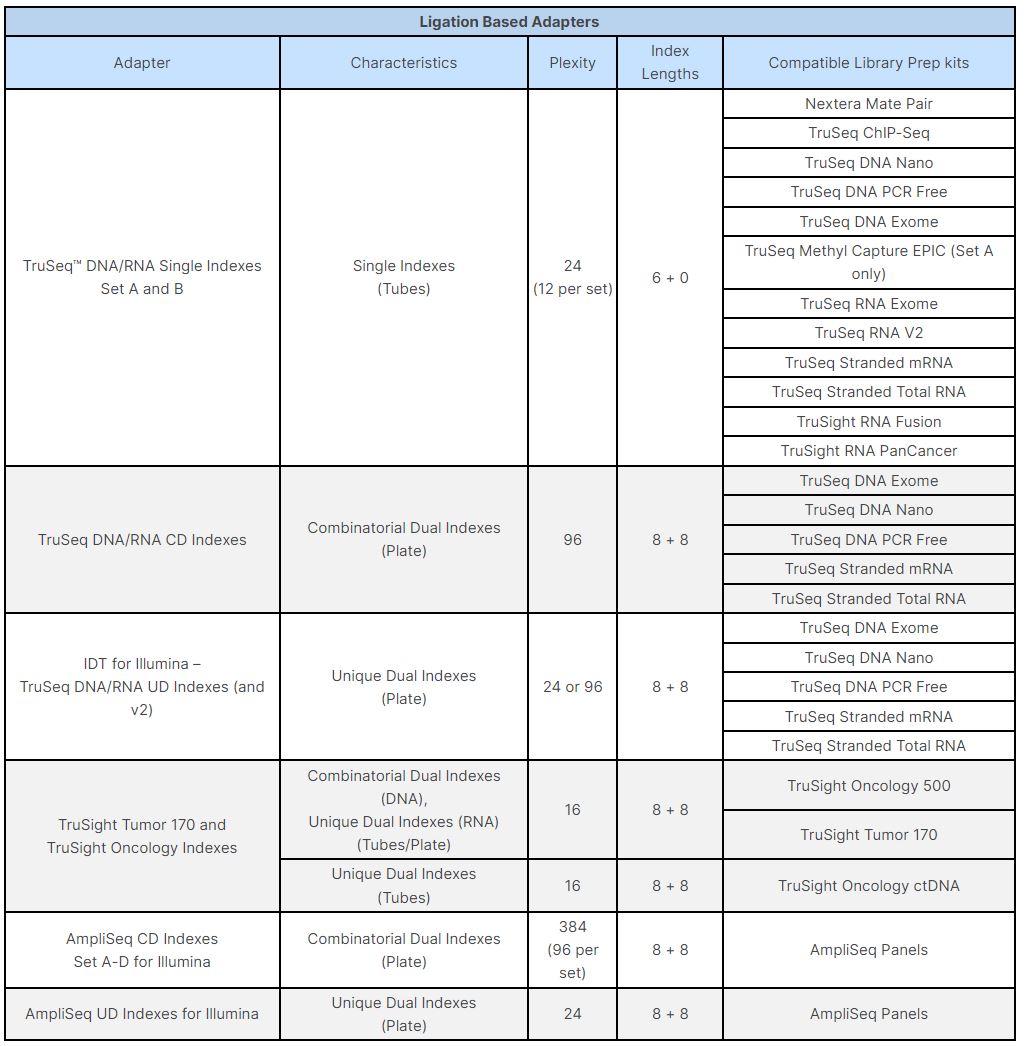
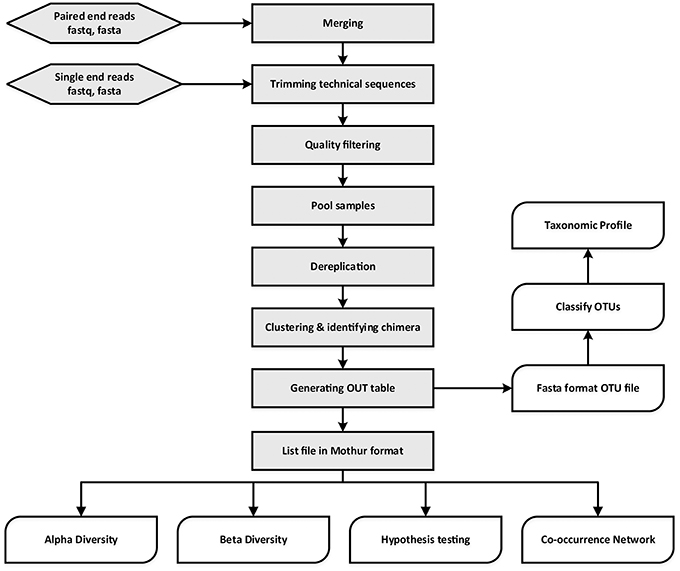
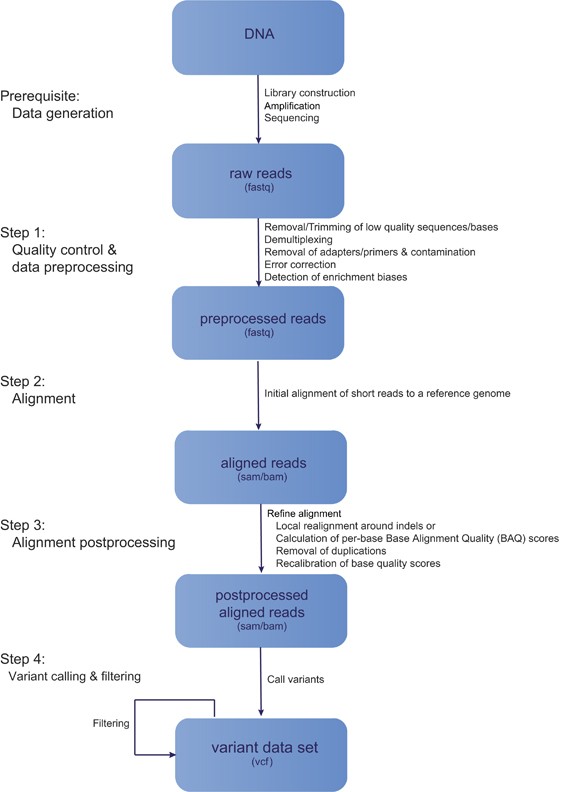
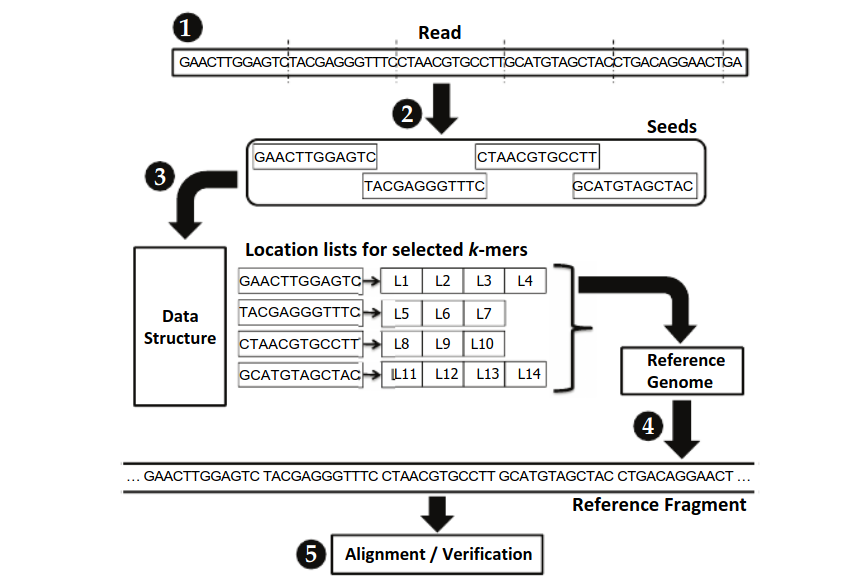
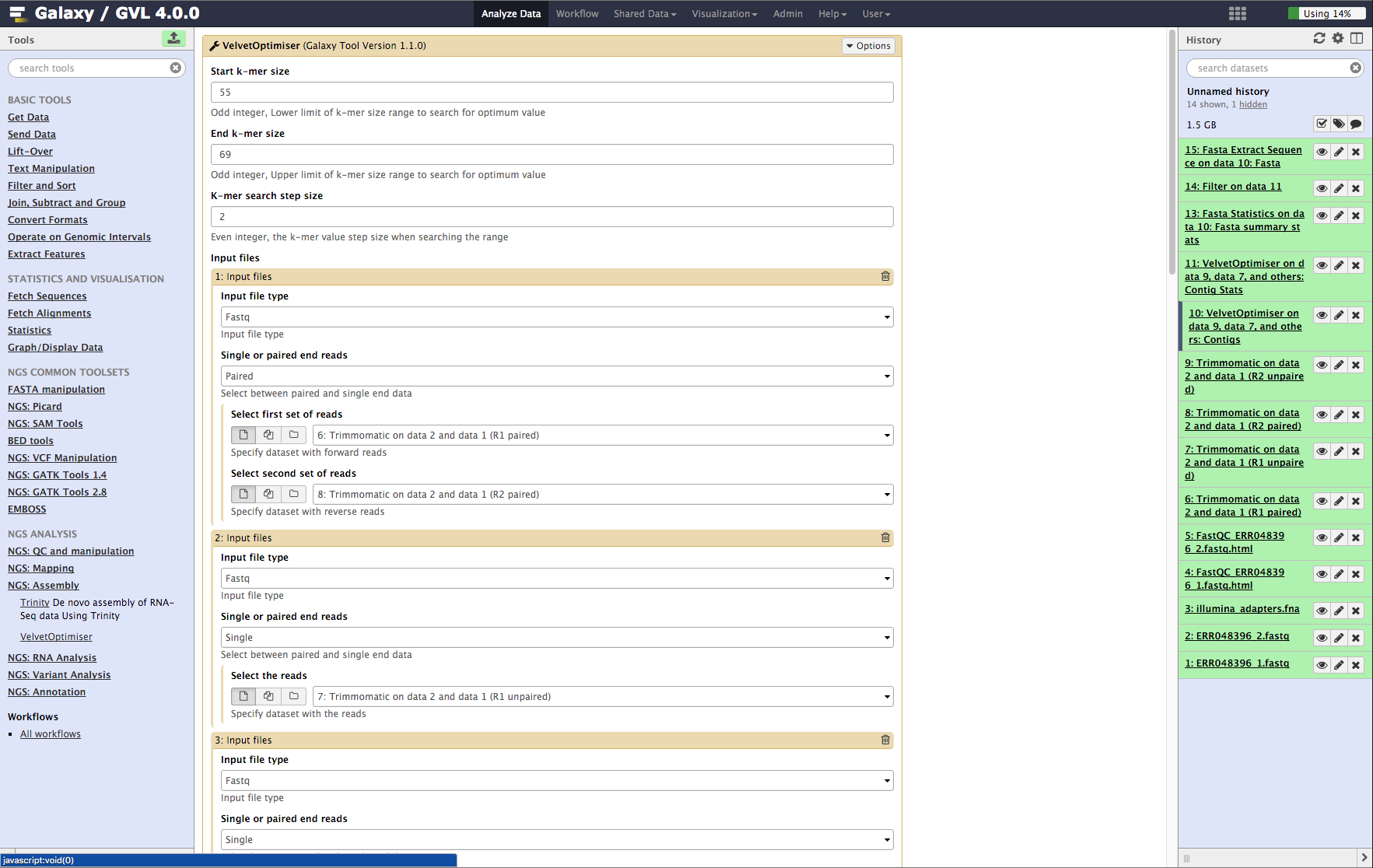
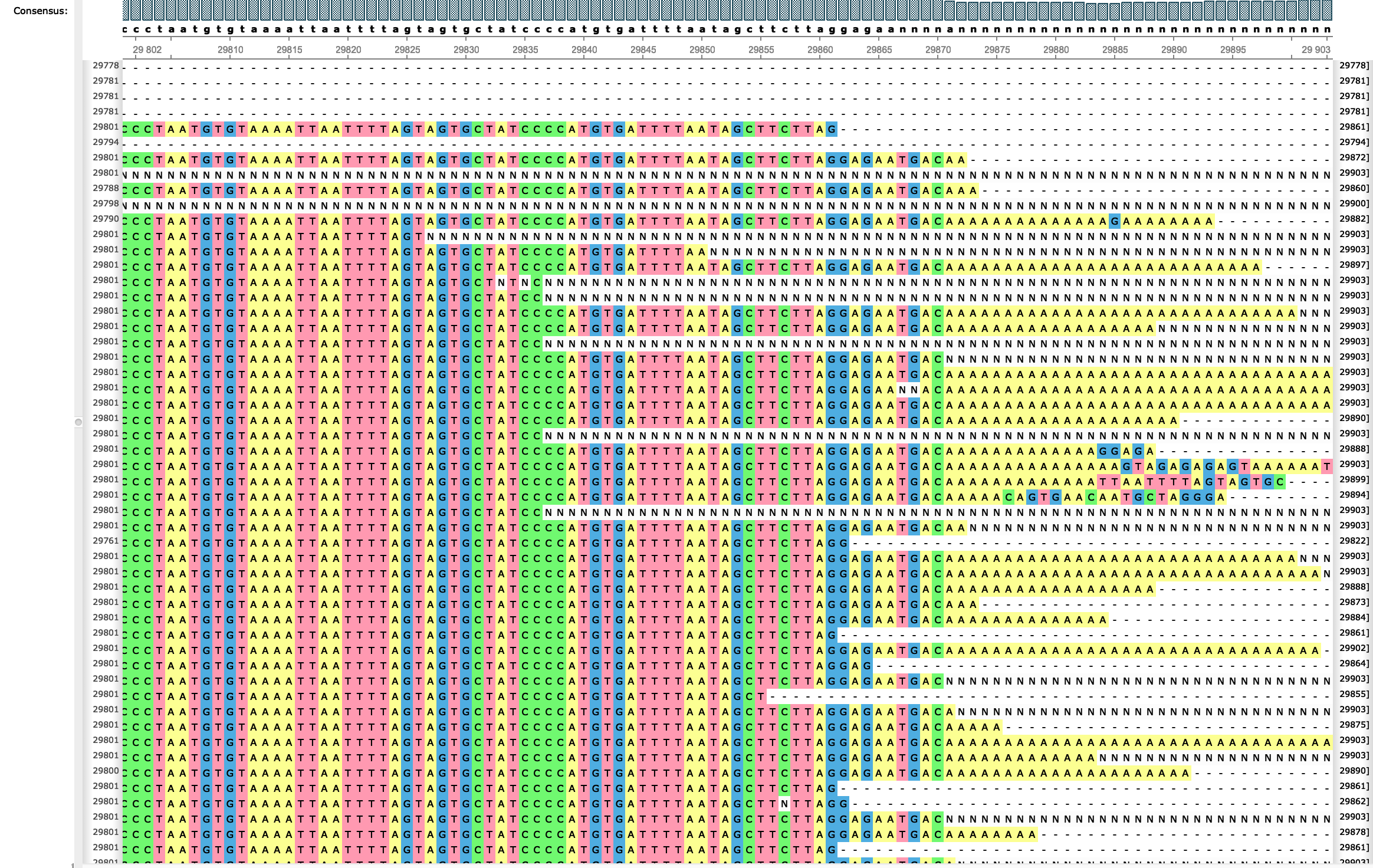
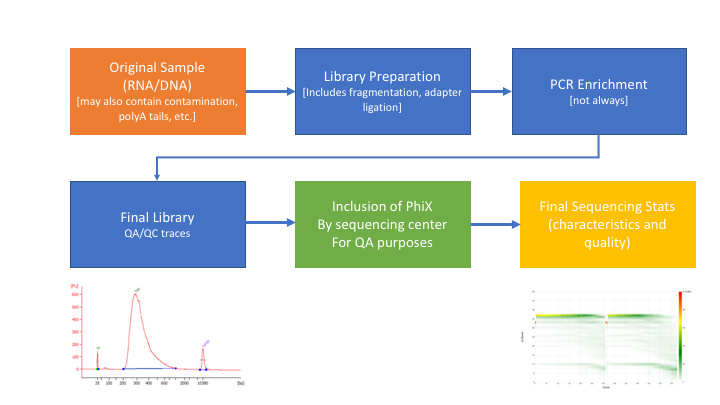
A single test approach for accurate and sensitive detection and taxonomic characterization of Trypanosomes by comprehensive anal
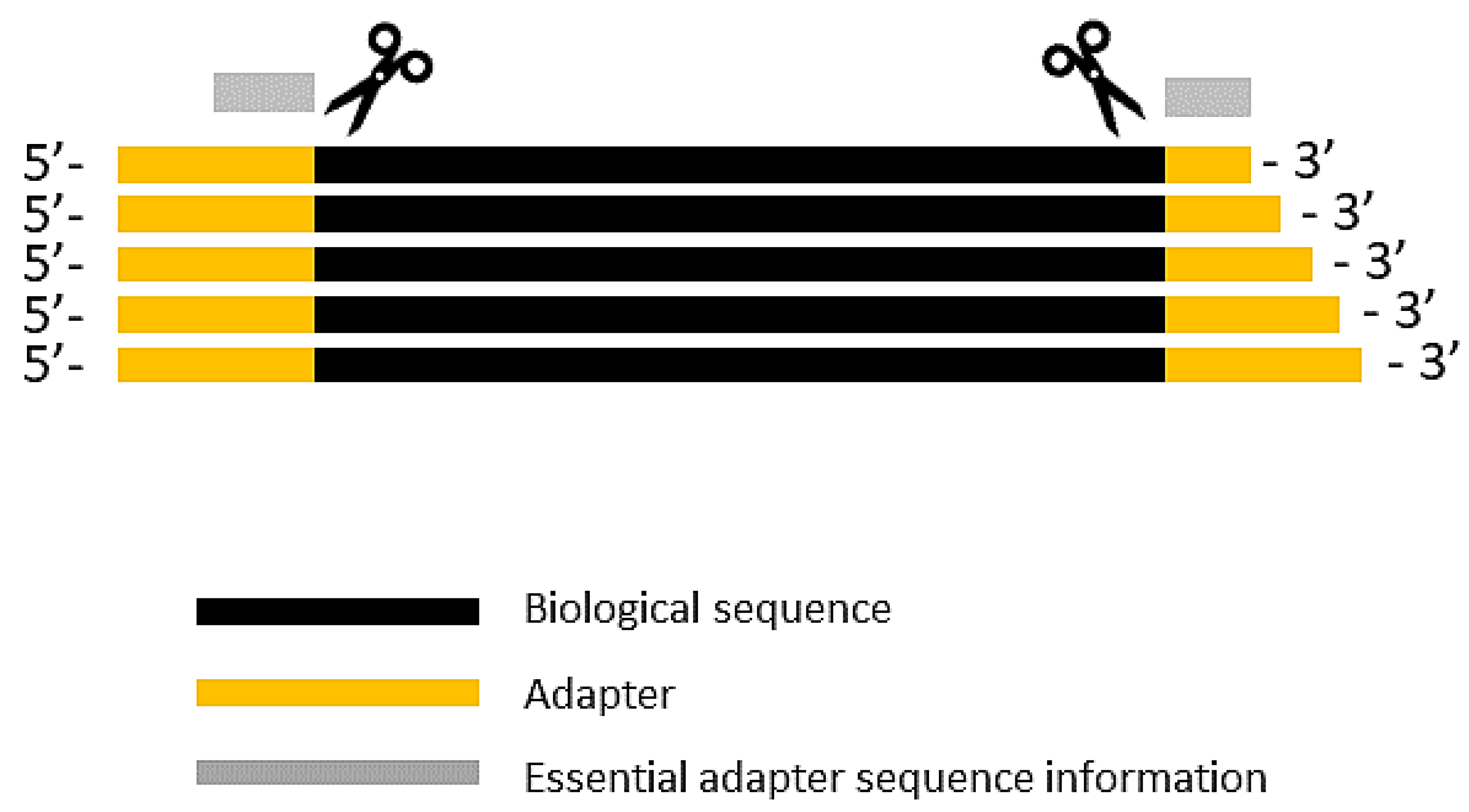
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

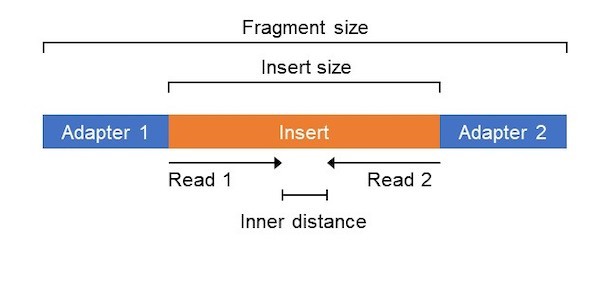
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge

FASTAptamer: A Bioinformatic Toolkit for High-throughput Sequence Analysis of Combinatorial Selections: Molecular Therapy - Nucleic Acids
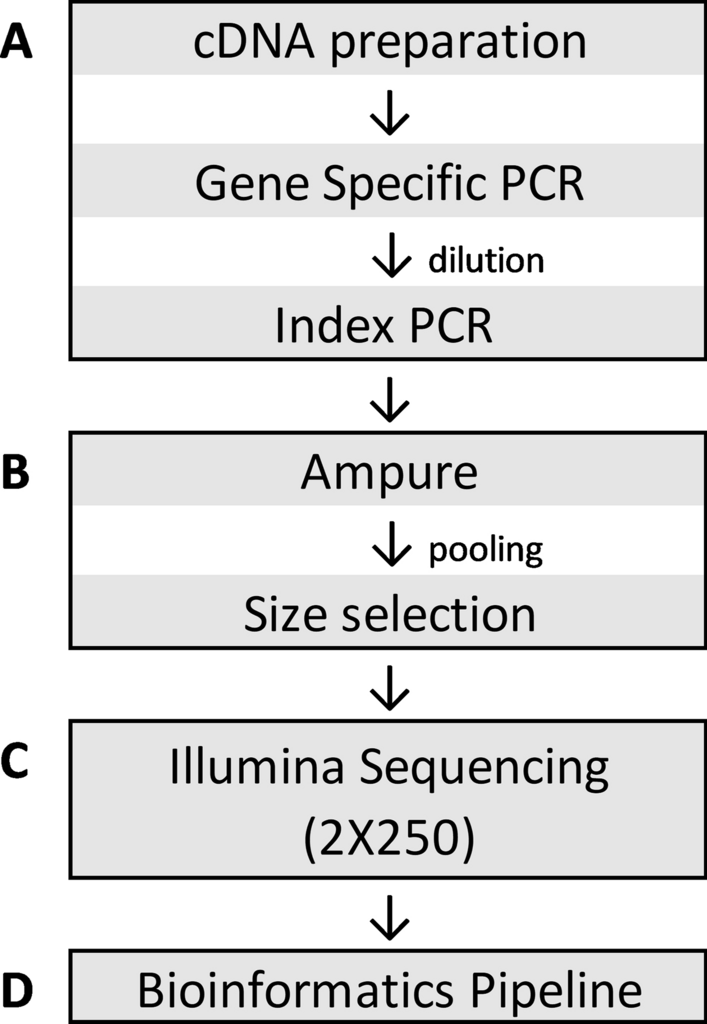
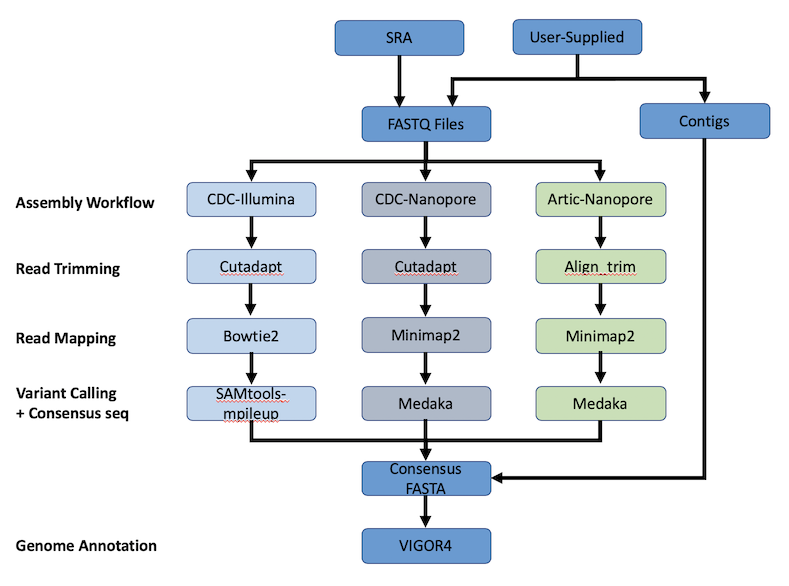
Development and validation of a high throughput SARS-CoV-2 whole genome sequencing workflow in a clinical laboratory | Scientific Reports

Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge
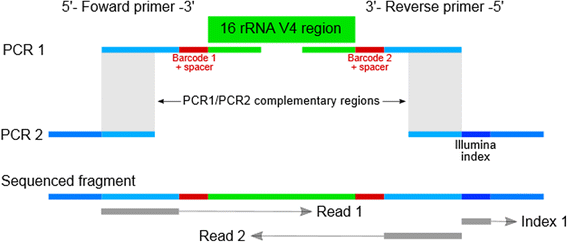
A novel ultra high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform | Microbiome | Full Text